Smallpox is a highly fatal human epidemic caused by the Variola virus (VAR), a member of the Orthopoxvirus genus of the Poxviridae family. The global eradication of smallpox in 1977 was made possible by the introduction of a highly safe and affordable vaccine derived from the Vaccinia virus (VAC), mass vaccination, and thorough epidemiological surveillance. After the eradication of smallpox, vaccination was discontinued worldwide. This provided an opportunity for zoonotic Orthopoxviruses, such as the Monkeypox virus (MPV), Cowpox virus (CPV), and Buffalopox virus, to spread and undergo adaptive mutations. MPV has garnered the most attention among zoonotic Orthopoxviruses. The clinical similarity between monkeypox and smallpox has sparked speculation about the genetic relationship between these viruses and whether MPV might evolve into a VAR-like virus with high-frequency human-to-human transmission.
Scientists from the State Research Center of Virology and Biotechnology "Vector" in Russia, including Sergei N. Shchelkunov, sequenced the 197 kb genome of MPV isolated from a patient during a large-scale monkeypox outbreak in Zaire in 1996. By comparing the genome sequence of MPV with those of the Indian 1967 strain of VAR (VAR-IND), which causes severe disease, and the relatively mild smallpox strain Garcia-1966 (VAR-GAR), they found that the nucleotide sequence in the central region of the MPV genome, which encodes essential enzymes and structural proteins, shares 96.3% homology with that of VAR. In contrast, the terminal regions encoding virulence and host range factors exhibit significant differences. The mutations in two interferon (IFN) resistance genes in MPV, as well as the presence of interleukin-1β (IL-1β) inhibitors, may account for the differences in characteristics between these two viruses and could also limit the use of MPV as a smallpox model. Although the extensive genetic differences confirm that MPV is not a direct "relative" of VAR, the possibility of future adaptation of MPV to humans cannot be ruled out.
The full-length genome of MPV-ZAI is 196,858 bp, containing 190 largely non-overlapping ORFs with ≥60 amino acid residues. Its structural features and GC content (31.1%) are similar to those of other Orthopoxviruses. The coding sequence of the MPV-ZAI gene is longer than that of VAR, at 195,118 bp. The longer length of MPV DNA is mainly due to the duplication of four terminal ORFs on the left side, which are part of the terminal inverted repeats (TIRs), while the VAR genome has very short non-genic TIRs lacking ORF duplication (Figure 1).
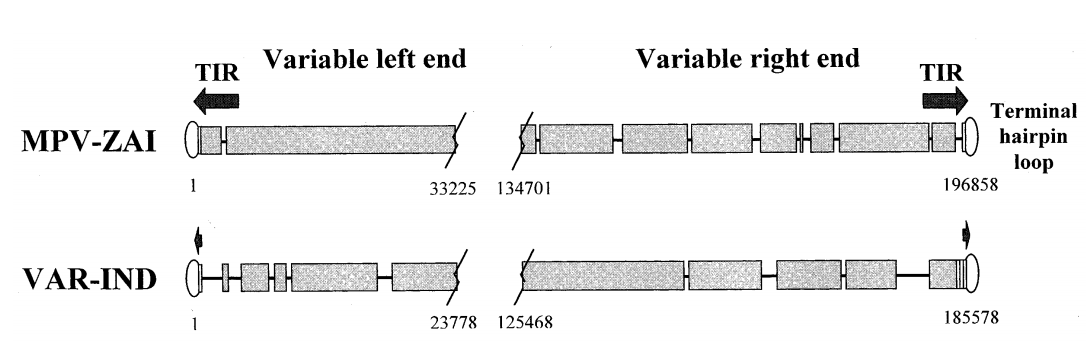
Figure 1. Schematic diagram of the terminal species-specific variable genome regions of MPV-ZAI and VAR-IND
Note: TIRs are indicated by arrows, and short tandem terminal repeat regions are shown as rectangles. Overlapping sequences are indicated by wide black bars; deletions in one genome relative to the other are shown as lines. The boundaries of variable genome regions are marked by nucleotide numbers corresponding to their positions in the genome.
The central genome region of Orthopoxviruses mainly contains highly conserved essential genes. The central genome region of MPV-ZAI DNA is 101,466 bp, defined by the C10L and A25R ORFs, with 96.3% homology to VAR-IND. The amino acid sequences of the virion proteins encoded in this region of MPV-ZAI show 91.7-99.2% similarity to those of VAR-IND.
Notably, mutations in the two IFN resistance genes of MPV-ZAI result in their translation into intracellular proteins, which are intact in VAR and other Orthopoxviruses. One of these proteins (C3L in VAR-IND, Table 1) is a homolog of eukaryotic translation initiation factor 2α (eIF-2α) and acts as a decoy to inhibit the antiviral activity of double-stranded RNA-dependent protein kinase (PKR). The MPV genomes (ZAI and CNG strains) do not encode this protein. Studies have shown that mutated VAC strains lacking this gene exhibit IFN sensitivity, with virus yields reduced by approximately 100-fold compared to the parent virus. Another IFN resistance gene (E3L in VAR-IND, Table 1) is present in VAR and other Orthopoxviruses, expressed as long or short forms depending on the first or second methionine (Figure 2A). In VAC, the long form's N-terminal domain mediates binding to Z-DNA, nuclear localization, and interaction with PKR, essential for VAC virulence. The C-terminal domain of both forms binds to double-stranded RNA, inhibiting IFN-induced PKR and 2-5A synthetase activation, which is necessary for IFN resistance and VAC host range. In MPV, the first translation start codon and downstream nonsense mutations result in only the short form being translated (Figure 2A). These two IFN mutations may contribute to the lower human-to-human transmission of MPV compared to VAR.
MPV encodes a complement-binding protein with only three short repeats, while other Orthopoxviruses have four (Figure 2B). Additionally, MPV encodes a secreted IL-1β-binding protein and 3-β-hydroxy-Δ5-steroid dehydrogenase, which have incomplete ORFs in VAR strains (Table 1). Notably, the VAC gene encoding the IL-1β-binding protein is associated with fever and pathogenicity. The presence of the IL-1β-binding protein in MPV may be one reason for its lower pathogenicity compared to VAR.
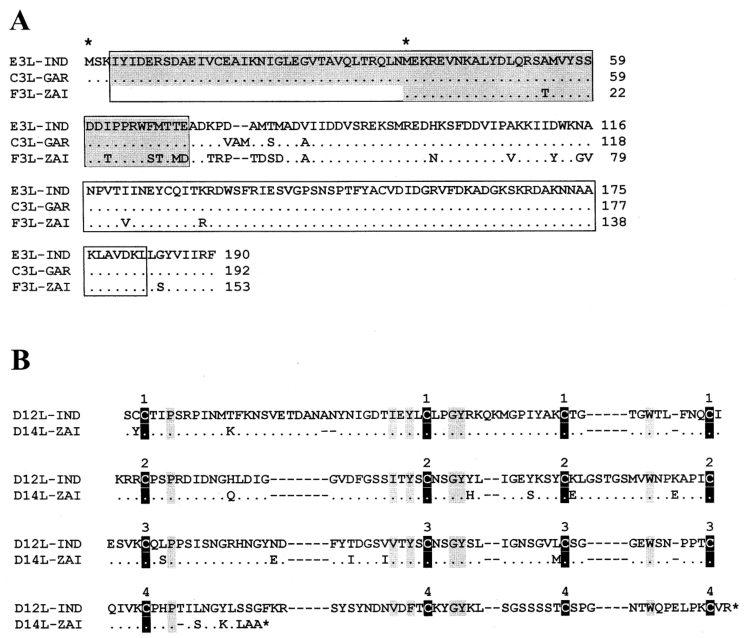
Figure 2. Alignment of amino acid sequences of IFN resistance factors and complement-binding proteins between MPV and VAR
*Notes: Figure 2A shows the alignment of amino acid sequences of the E3L IFN resistance factors encoded by corresponding genes in VAR-IND, VAR-GAR, and MPV-ZAI. Regions containing the N-terminal adenosine deaminase Z-α domain (gray) and C-terminal double-stranded RNA-binding motifs are indicated. Identical amino acid residues are marked with dots; deletions are marked with dashes. The first and second methionine residues starting the long or short forms of the protein are indicated with asterisks above the sequence. Figure 2B shows the alignment of complement-binding protein amino acid sequences in VAR-IND and MPV-ZAI ORFs. Conserved cysteine residues are indicated by black vertical bars, and other conserved residues are indicated by gray vertical bars. The numbers above the boxes represent the four typical repeats of the complement control protein.
Table 1. Comparison of virulence factors between MPV and VAR
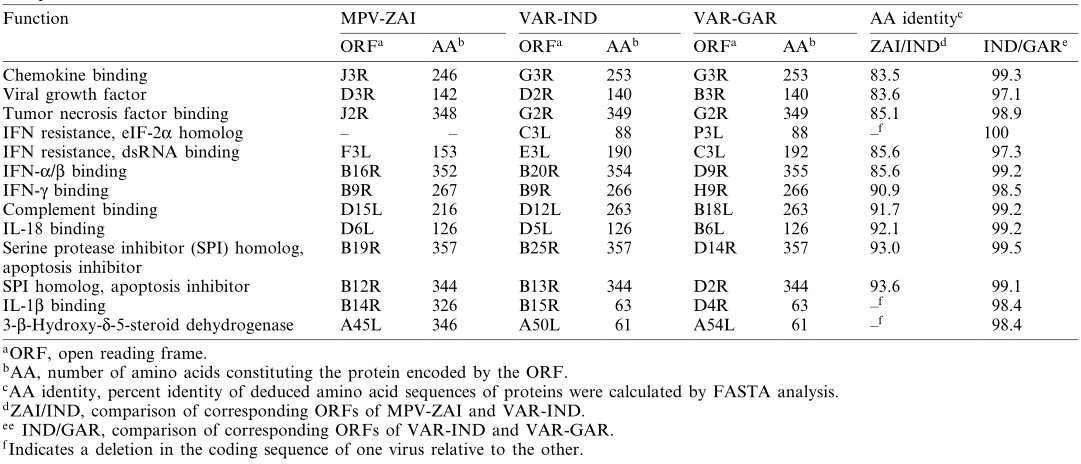
ORFs encoding ankyrin repeat-containing proteins, some of which have host range functions (MPV-ZAI D7L and C1L, homologous to Orthopoxvirus), form the largest gene family in Orthopoxviruses. Among the 10 genes in this family, the gene corresponding to VAR-IND B19R is deleted in the MPV-ZAI genome, and the gene corresponding to D1L in MPV-ZAI is missing in both VAR strains. Additionally, four VAR genes in this family (D6L, D7L, C1L, and O3L in VAR-IND) are truncated compared to their MPV homologs. Currently, we can only speculate on how these differences might affect host range or virulence.
Table 2. Comparison of ankyrin repeat proteins between MPV and VAR

To better understand the genetic relationships, a phylogenetic analysis of the terminal variable genome regions of four human pathogenic Orthopoxviruses was performed using 117,600 bp of DNA (Figure 3). The major and minor subspecies of VAR are closely related, and MPV is slightly more distant from VAR than VAC.
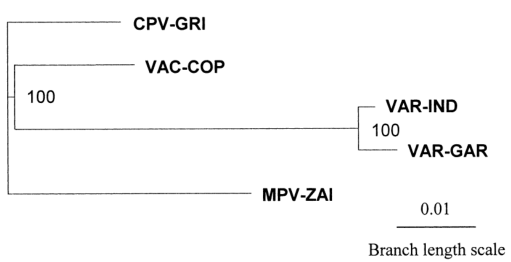
Figure 3. Phylogenetic tree analysis of the terminal variable genome sequences of MPV, VAR, CPV, and VAC
In conclusion, the comparison of the genomes of MPV and VAR reveals multiple differences in virulence genes between the major and minor strains of VAR. MPV and VAR likely evolved independently from a common Orthopoxvirus ancestor. The genetic differences between VAR and MPV raise significant questions about the validity of using MPV as a smallpox model. However, given the severe diseases caused by MPV, close monitoring of MPV infection rates is essential to ensure that it does not undergo human adaptation, either spontaneously or through recombination, especially in unvaccinated populations with high HIV prevalence.
References
1.Shchelkunov S.N. et al. (2001). Analysis of the Monkeypox Virus Genome. Virology Journal.
 亚洲精品无码久久久久
|
亚洲日本日韩中文字幕狼友版
|
国产精品高潮久久久久无码
|
免费观看成人久久网免费观看
|
国产精品亚洲专区在线播放
|
91亚洲一区二区三区
|
亚日韩一二三区视频免费看
|
精品一级 片内射视网站
|
亚洲精品日韩精品一乛方
|
我把护士日出水了视频90分钟
|
亚洲尤码不卡AV麻豆
|
国产伦精品一区二区三区免费观看
|
欧美日韩无线码一区茄子视频
|
人人狠久久88全国最大色
|
肉色超薄丝袜脚交一区二区
|
亚洲自偷自拍熟女另类
|
老司机久久精品最新免费
|
免费又黄又硬又爽大片免费
|
久久久久成亚洲国产av综合精品
|
内射中出无码护士在线
|
久久综合精品国产一区无码
|
国内2021自在自线
|
欧美又大又硬又粗BBBBB
|
91酒店疯狂输出女神范范
|
精品视频无码一区二区三区
|
99国产精品一区二区三区四区五区
|
国产精品991TV制片厂在线观看
|
国产亚洲日韩欧美另类第八页
|
亚洲AV无码乱码在线观看侵犯
|
国产国语**毛片高清视频
|
美女黄色视频在线观看
|
亚洲宅男精品一区在线观看
|
扒开双腿猛进入爽爽在线观看
|
综合激情区视频一区视频二区
|
国语A在线看免费观看视频
我和闺蜜在公车被cao污文
|
亚洲无码啪啪啪啪啪啪
|
国产JIZZJIZZ免费看
|
国产高清av首播原创麻豆
|
久久精品99久久久久久蜜芽TV
|
国产精品内射后入合集
|
秋葵视频ios无限制解码免费版下载
|
亚洲精品无码久久久久
|
亚洲日本日韩中文字幕狼友版
|
国产精品高潮久久久久无码
|
免费观看成人久久网免费观看
|
国产精品亚洲专区在线播放
|
91亚洲一区二区三区
|
亚日韩一二三区视频免费看
|
精品一级 片内射视网站
|
亚洲精品日韩精品一乛方
|
我把护士日出水了视频90分钟
|
亚洲尤码不卡AV麻豆
|
国产伦精品一区二区三区免费观看
|
欧美日韩无线码一区茄子视频
|
人人狠久久88全国最大色
|
肉色超薄丝袜脚交一区二区
|
亚洲自偷自拍熟女另类
|
老司机久久精品最新免费
|
免费又黄又硬又爽大片免费
|
久久久久成亚洲国产av综合精品
|
内射中出无码护士在线
|
久久综合精品国产一区无码
|
国内2021自在自线
|
欧美又大又硬又粗BBBBB
|
91酒店疯狂输出女神范范
|
精品视频无码一区二区三区
|
99国产精品一区二区三区四区五区
|
国产精品991TV制片厂在线观看
|
国产亚洲日韩欧美另类第八页
|
亚洲AV无码乱码在线观看侵犯
|
国产国语**毛片高清视频
|
美女黄色视频在线观看
|
亚洲宅男精品一区在线观看
|
扒开双腿猛进入爽爽在线观看
|
综合激情区视频一区视频二区
|
国语A在线看免费观看视频
我和闺蜜在公车被cao污文
|
亚洲无码啪啪啪啪啪啪
|
国产JIZZJIZZ免费看
|
国产高清av首播原创麻豆
|
久久精品99久久久久久蜜芽TV
|
国产精品内射后入合集
|
秋葵视频ios无限制解码免费版下载
|